- Home
- About Us
- Students
- Academics
-
Faculty
- Electrical Engineering
- Automation
- Computer Science & Engineering
- Electronic Engineering
- Instrument Science and Engineering
- Micro-Nano Electronics
- School of Software
- Academy of Information Technology and Electrical Engineering
- School of Cyber Security
- Electrical and Electronic Experimental Teaching Center
- Center for Advanced Electronic Materials and Devices
- Cooperative Medianet Innovation Center
- Alumni
-
Positions
-
Forum
News
- · Bin Dai's Team Unveils the Assembly Mechanism of β-Lactoglobulin Fibrils, Providing New Insights for the Development of Functional Nanomaterials
- · Mingyi Chen’s research group has made important progress in the field of analog-to-digital converter chips for brain-computer interface
- · Progress in the Development of Semiconductor Nanomaterials to Activate Pyroptosis for Cancer Therapy
- · Jiamiao Yang’s team achieved the high precision optoelectronic reservoir computing based on complex-value encoding
- · Significant Advancements in Resonator-Enhanced Quantum Sensing Achieved by Zenguihua's Team at the School of Sensing Science and Engineering
Shanghai Jiao Tong University's Shen Hongbin and Yuan Ye team have made new advances in bioinformatics research on transcriptional regulation RNA velocity estimation.
Recently, the team of Shen Hongbin and Yuan Ye from the School of Electronic Information and Electrical Engineering at Shanghai Jiao Tong University published a research paper titled 'TFvelo: gene regulation inspired RNA velocity estimation' in Nature Communications. They proposed a novel computational method called TFvelo for RNA velocity modeling, inspired by gene regulation at the transcriptional level.

In recent years, RNA velocity has emerged as a highly researched direction in the field of single-cell transcriptomic analysis. It involves establishing dynamic equations to simulate the changes in RNA expression levels, thereby enabling inference of complex cellular trajectories. Previous modeling of RNA velocity often relied on signals from single RNA splicing, resulting in less accurate predictions of gene dynamics inferred from high-noise data. By analyzing the timescale of splicing processes and the intensity of noise in single-cell data, the authors found that signals from individual genes were insufficient to accurately fit the dynamic changes in gene expression. As a result, they proposed the RNA velocity modeling method TFvelo based on the transcriptional regulatory relationships between multiple genes. By utilizing the Generalized EM algorithm for model optimization, they improved the accuracy of modeling gene dynamics and cell differentiation trajectories.
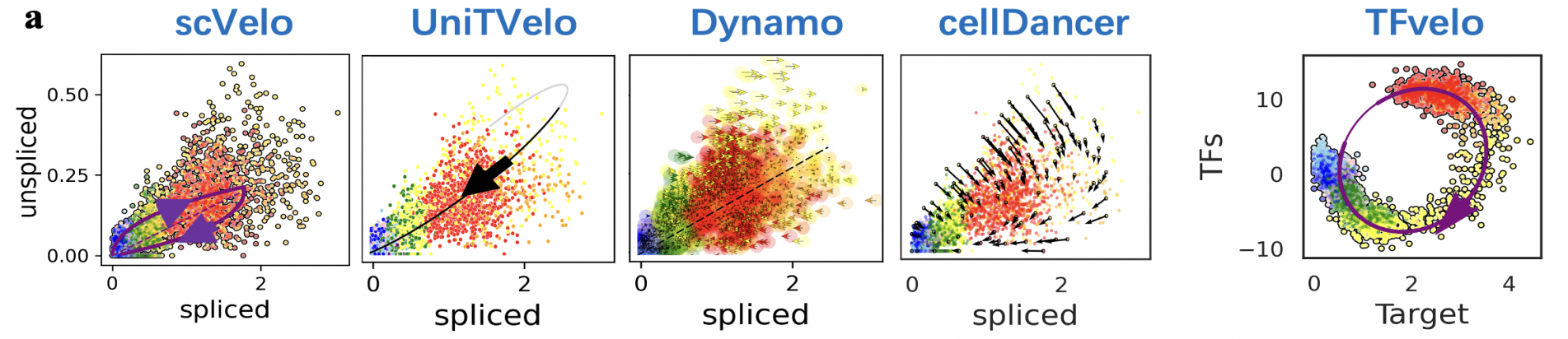
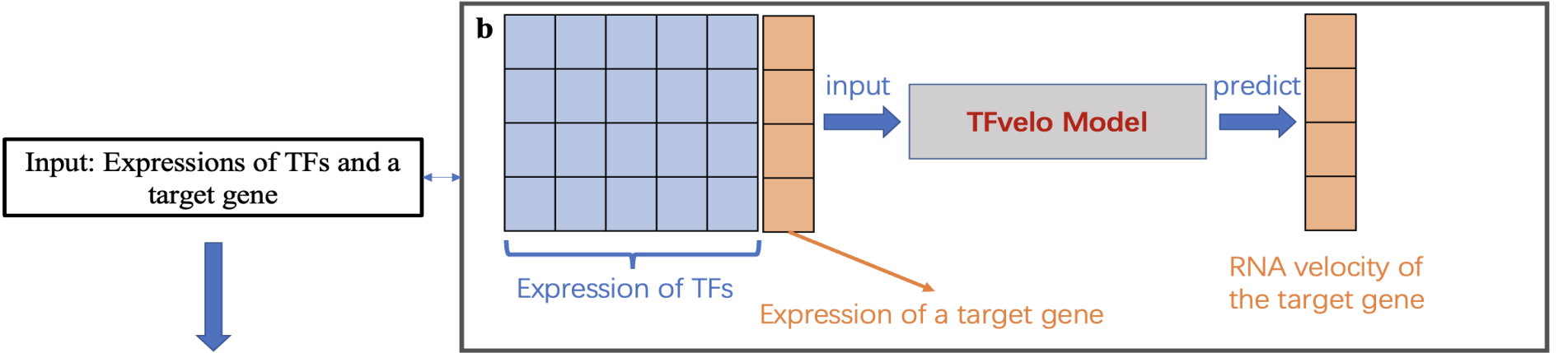
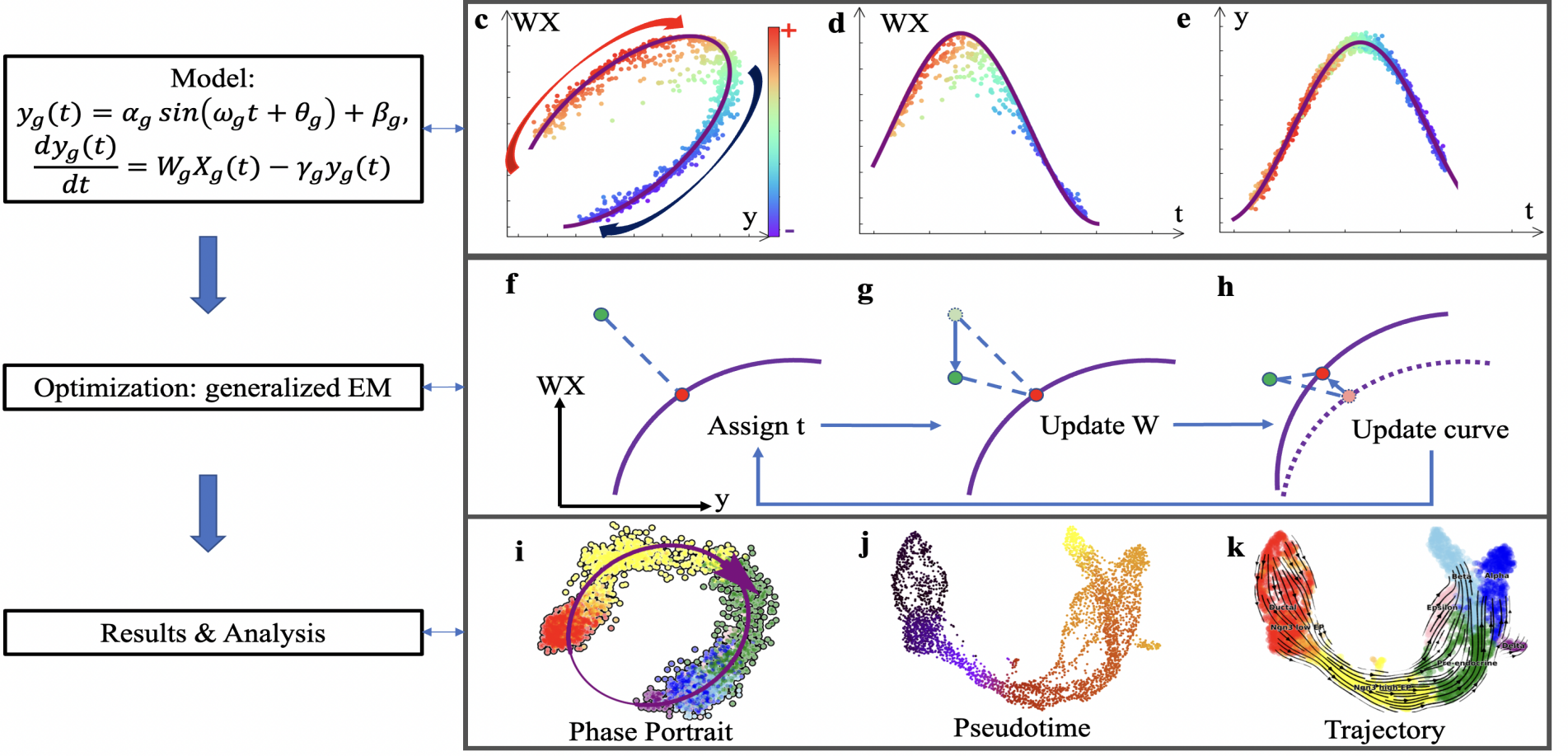
Model Flowchart
Meanwhile, because TFvelo utilizes information from multiple transcription factors, it can learn the weights corresponding to each transcription factor from the data and their phase differences in changes with different genes. Consequently, it can further analyze the transcriptional regulatory relationships between genes. TFvelo can also extend the RNA velocity model to datasets without splicing information annotations, thus expanding its applicability to a wider range of scenarios. TFvelo provides new modeling approaches and methods for single-cell trajectory analysis, holding potential application value in exploring mechanisms such as cell differentiation, tissue development, and tumor evolution.
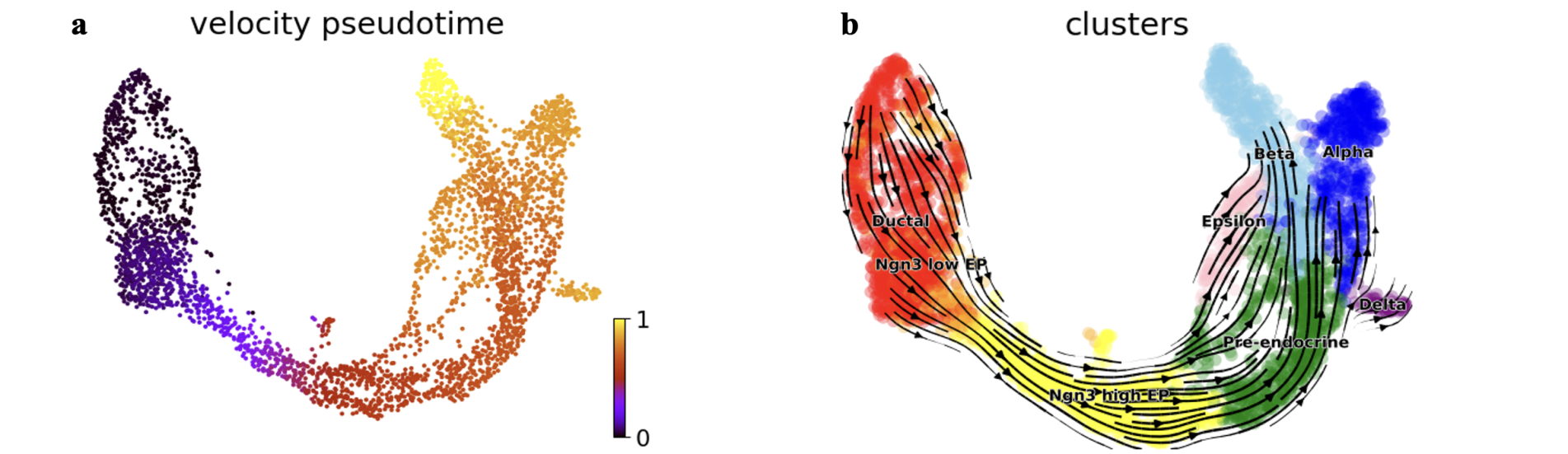
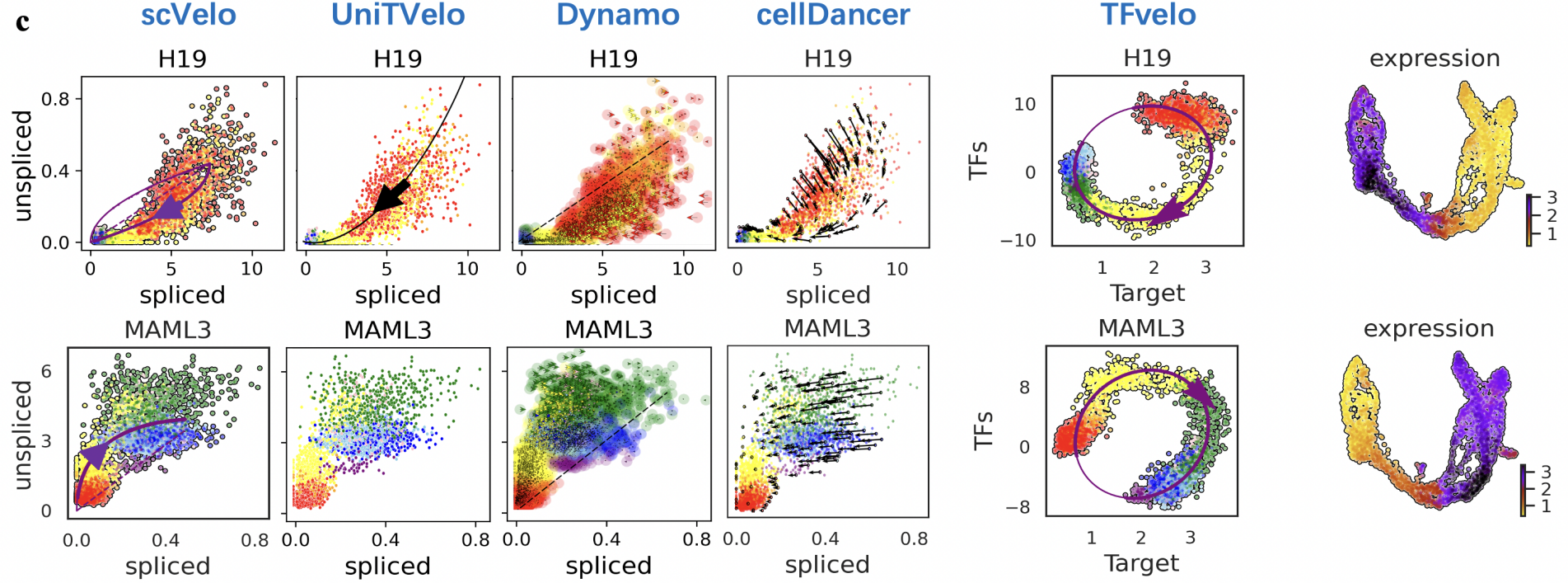
The predictive accuracy of TFvelo for cell trajectories and its fitting effectiveness for single-gene dynamic processes
Doctoral candidate Li Jiachen from the Department of Automation is the first author of the paper, with Associate Professor Yuan Ye and Professor Shen Hongbin from the Department of Automation as corresponding authors.
-
Students
-
Faculty/Staff
-
Alumni
-
Vistors
-
Quick Links
